About me
Currently, I’m working as a Scientific Collaborator at ETH Zurich on further scaling up MetaGraph, a large-scale sequence search engine for DNA, and I’m open to new projects.
Previously, I was a Machine Learning researcher and engineer at Kaiko.AI, where I trained Vision Transformers (ViTs) and built foundation models (FMs) for computational pathology and other data modalities. This includes large-scale pretraining on diverse datasets, fine-tuning and optimizing the models for various downstream tasks, and making these models robust, interpretable, and useful in clinical practice.
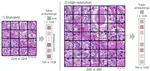
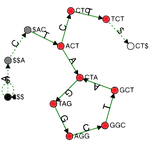
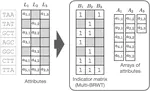
I completed my PhD at ETH Zurich, where I designed novel algorithms and compressed data structures for indexing petabytes of biological sequences and developed methods scalable to the entire Sequence Read Archive. These methods finally made this trove of data accessible for search by sequence at scale and demonstrated the feasibility of making all of life's code easily searchable.
Before that, I studied Math, Physics, CS, and Machine Learning at MIPT, Skoltech, and YSDA and worked on a few problems of computational structural biology at Inria Grenoble-Rhône-Alpes.
- Machine Learning
- Bioinformatics
- Computational Biology
- Compressed Data Structures
- Hiking/Camping, Skiing, Biking
- Guitar, Piano
-
Ph.D. in Computer Science, 2023
ETH Zurich, Zurich, Switzerland
-
M.Sc. in Math. and Computer Science, 2017
Skoltech, Moscow, Russia
-
M.Sc. in Applied Math. and Physics, 2017
MIPT, Moscow, Russia
-
PG Dip. in Computer Science, 2016
Yandex School of Data Analysis, Moscow, Russia
-
B.Sc. in Applied Math. and Physics, 2015
MIPT, Moscow, Russia
Featured Projects
Featured Publications
Events & Talks
Teaching
Courses TAed at ETH Zürich, Institute for Machine Learning:
-
Advanced Machine Learning (Fall 2020, 2019, 2018)
-
Computational Challenges in Medical Genomics (Spring 2022, 2021, 2020, 2019)
-
Computational Intelligence Lab (Spring 2019, 2018)
-
Deep Learning (Fall 2017)
-
Introduction to Machine Learning (Spring 2020)
-
Statistical Learning Theory (Spring 2021)