Abstract
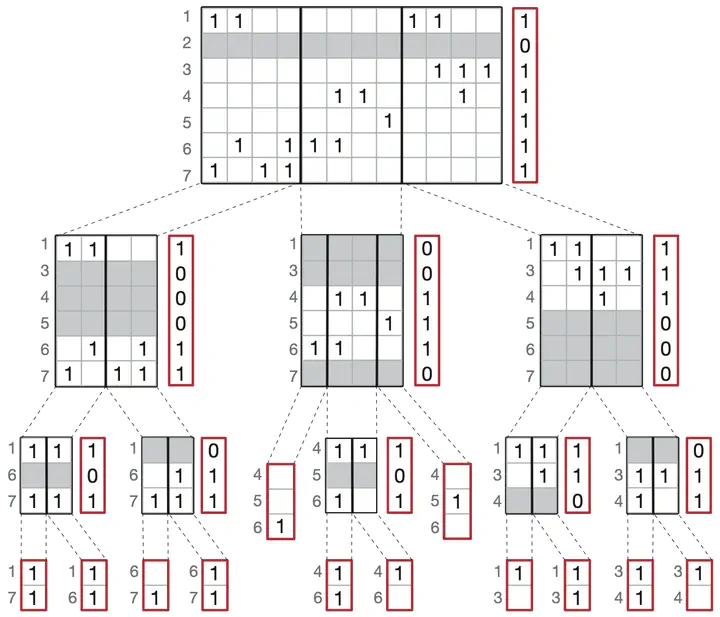
High-throughput DNA sequencing data are accumulating in public repositories, and efficient approaches for storing and indexing such data are in high demand. In recent research, several graph data structures have been proposed to represent large sets of sequencing data and to allow for efficient querying of sequences. In particular, the concept of labeled de Bruijn graphs has been explored by several groups. Although there has been good progress toward representing the sequence graph in small space, methods for storing a set of labels on top of such graphs are still not sufficiently explored. It is also currently not clear how characteristics of the input data, such as the sparsity and correlations of labels, can help to inform the choice of method to compress the graph labeling. In this study, we present a new compression approach, Multi-binary relation wavelet tree (BRWT), which is adaptive to different kinds of input data. We show an up to 29% improvement in compression performance over the basic BRWT method, and up to a 68% improvement over the current state-of-the-art for de Bruijn graph label compression. To put our results into perspective, we present a systematic analysis of five different state-of-the-art annotation compression schemes, evaluate key metrics on both artificial and real-world data, and discuss how different data characteristics influence the compression performance. We show that the improvements of our new method can be robustly reproduced for different representative real-world data sets.