Abstract
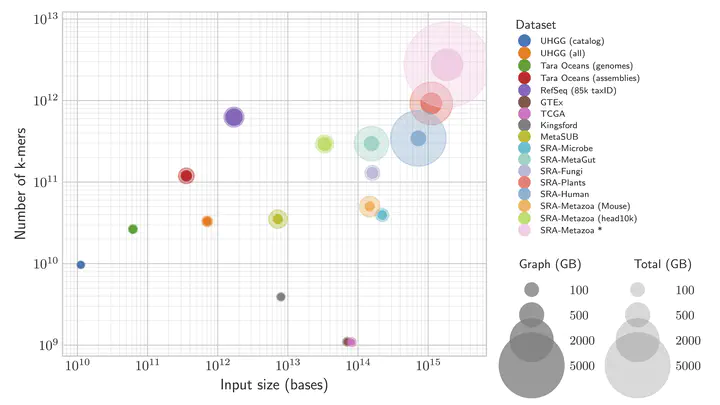
Technological advances have led to an exponential growth in the amount of high-throughput sequencing data available to the scientific community. However, most of this data, commonly stored in repositories such as the NCBI Sequence Read Archive, which currently contains over 50 petabases of sequences, is only available in raw format. Transforming it into a searchable representation that is easily accessible to life science researchers for large scale analysis and search remains an unsolved problem. In my talk, I will review the state-of-the-art approaches for indexing large cohorts of sequencing data. Then, I will describe MetaGraph, a method that efficiently indexes petabase-scale cohorts of sequencing experiments in annotated de Bruijn graphs and that supports k-mer search and sequence-to-graph alignment. Internally, MetaGraph represents input data as collections of k-mer sets encoded in succinct data structures, offering practically relevant trade-offs between the index size and the query performance. This flexibility allows running MetaGraph at different scales and on different hardware, from laptops to research compute clusters and distributed cloud environments. I will pay special attention to the methods and data structures used in MetaGraph for representing graph annotations, including non-binary attributes with Counting de Bruijn graphs, e.g., for representing gene expression and genome coordinates. Finally, I will conclude with real-world applications, such as indexing a portion of all publicly available whole-genome sequencing samples from the Sequence Read Archive, currently including over 90% of all Microbe, Fungi, Plant, and Human, as well as indexing all reference genome sequences (RefSeq), the RNA-Seq Genotype-Tissue Expression dataset (GTEx), and significantly more diverse metagenomic data, such as the entire catalog of 286,997 reference genome sequences from the human gut microbiome (UHGG), all 242,619 publicly available human gut microbiome short read sequencing samples, and a set of 4,220 public transit surface microbiomes (MetaSUB).