Projects
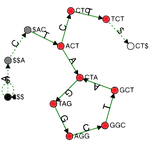
A C++ framework library for indexing very large collections of DNA/Protein sequences and a tool for sequence search, alignment, and assembly. Although the target use cases of MetaGraph overlap with BLAST, MetaGraph mainly focuses on the scalable indexing of raw sequencing data in annotated de Bruijn graphs with up to $\sim 10^{12}$ nodes and $\sim 10^{7}$ annotation labels. It also provides an online platform MetaGraph Online. Other contributors: Marc Zimmermann, Thomas Zhou, Oleksandr Kulkov, and the MetaGraph team.
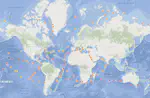
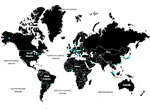
A portal for sequence search and geographical positioning based on the metagenomic MetaSUB data. The initial prototype was set up on a weekend but it served well and was also used as a base for the MetaGraph Search platform. Other contributors: Marc Zimmermann, Jiayu Chen, André Kahles, Thomas Zhou. (Published in Cell).